|
||
|
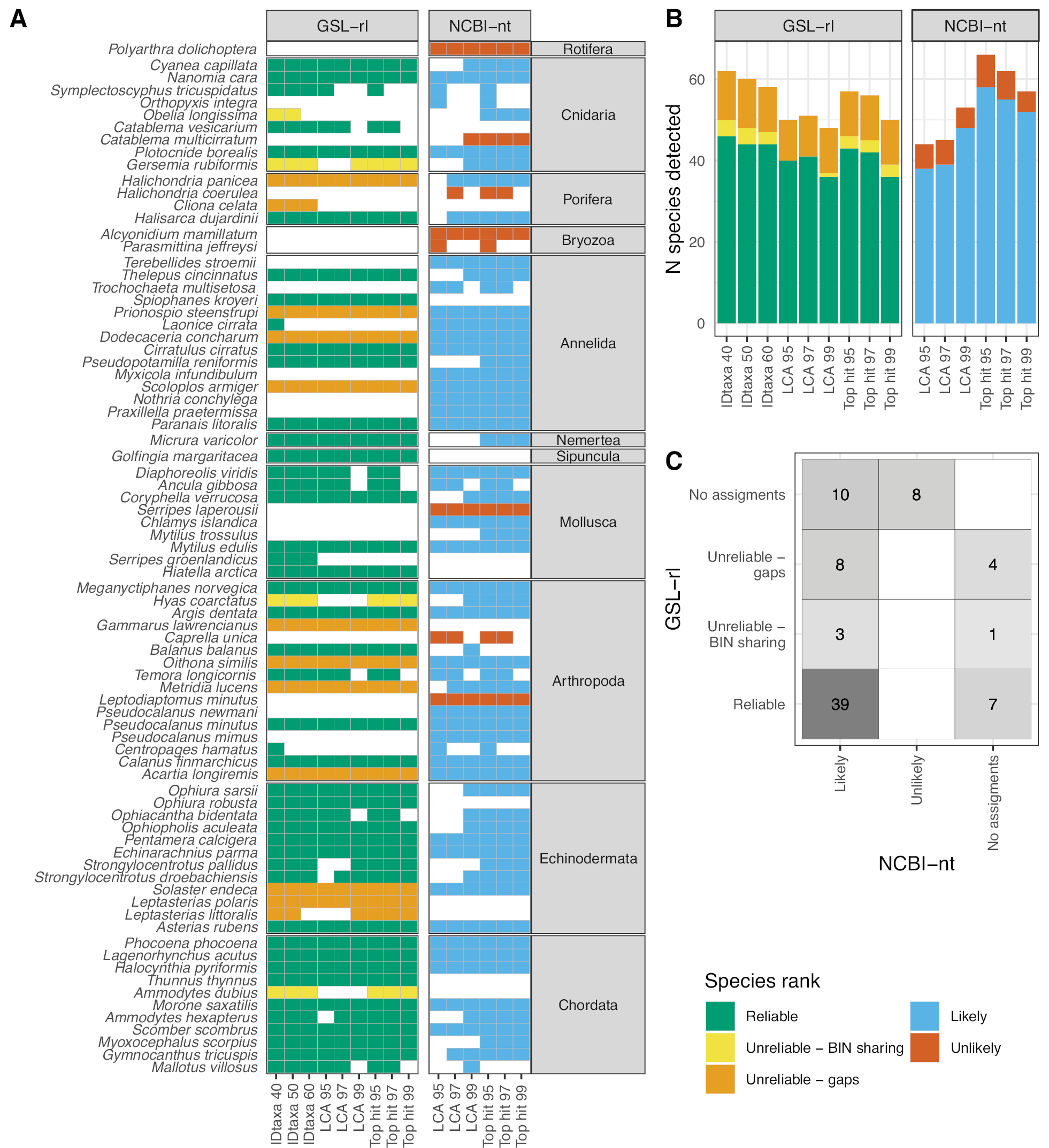
Assignment results at the species level using a regional library (GSL-rl) or a public repository (NBCI-nt) and popular assignment methods. We used three assignment methods, namely IDtaxa (confidence levels: 40%, 50% and 60%), BLAST-LCA, and BLAST-TopHit (identity thresholds: 95%, 97% and 99%). A Detections for each species and B number of species assignments for each source of reference sequences and method; C Comparison of species rank for all the species assigned with the two sources. Species rank categories are based on sequence availability and sequence similarity to closely related species in the Gulf of St. Lawrence for GSL-rl and on the geographic plausibility for NCBI-nt. |