|
||
|
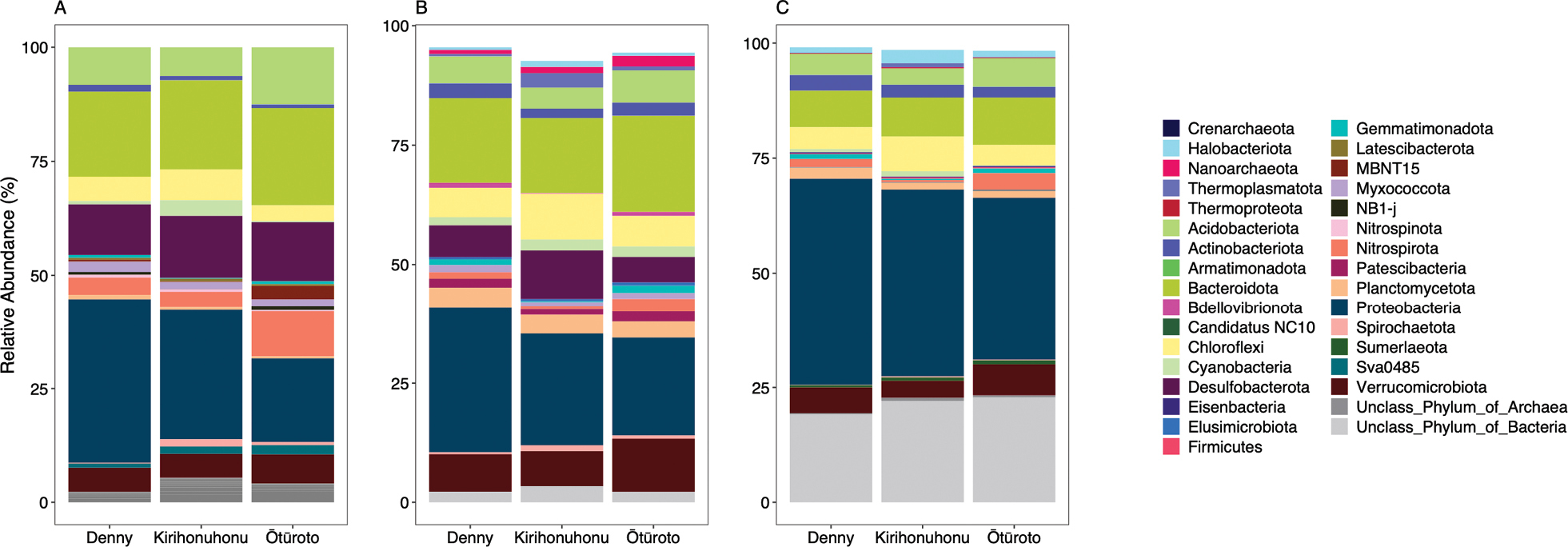
Taxonomic composition of the three Lakes at the phyla level for; A) 16S rRNA gene metabarcoding, B) 16S rRNA genes from the metagenomics and C) coding sequences from the metagenomics data. Only the top 20 phyla (based on mean abundance) are plotted for each method. |