|
||
|
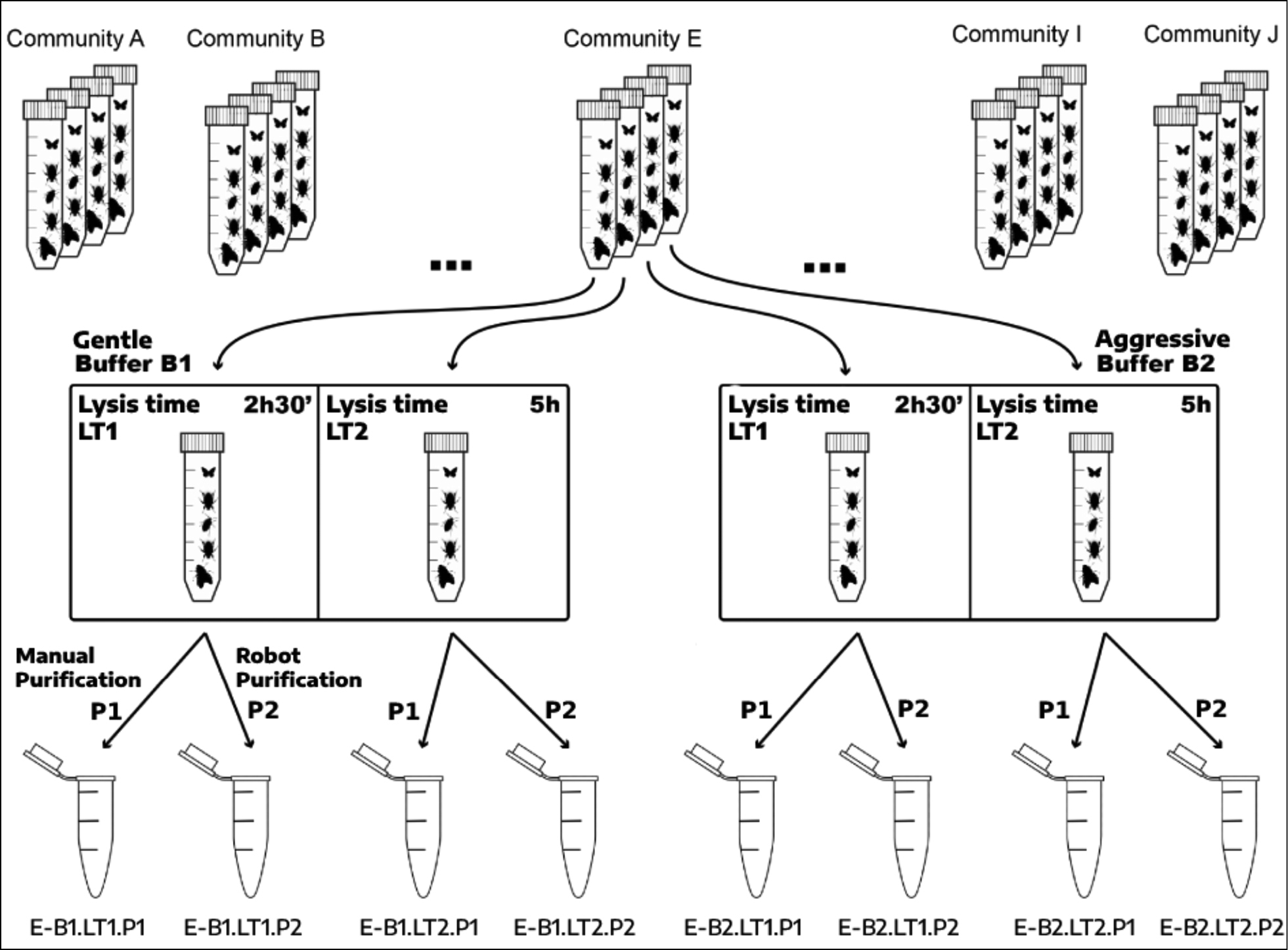
Schematic overview of the experimental design and treatments. Each community (A–J) was represented by four initial replicates (four tubes containing the same mix of species, each of them with the same number of specimens). Each replicate was incubated in one buffer (B1 or B2) for 2 h 30’ (LT1) or 5 h (LT2). Then, DNA from each lysate (B1–LT1, B1–LT2, etc.) was extracted by using both a manual salt saturation-salt protocol (P1) or silica-coated magnetic beads in a robot (P2). Thus, for each original community (A–J), eight DNA extracts were obtained, one from each combination of purification method, buffer and lysis time. |