|
||
|
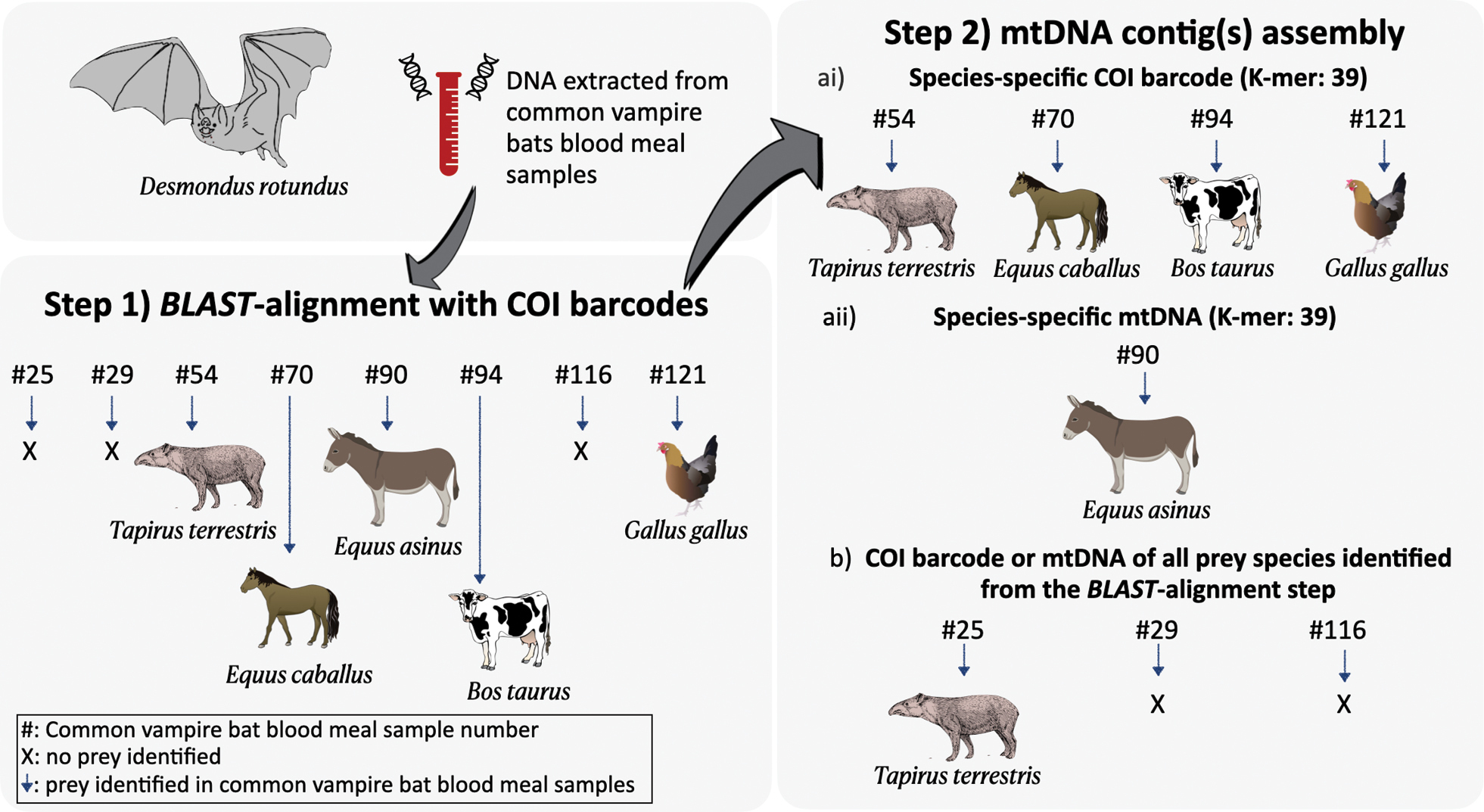
Prey identification of common vampire bat (Desmodus rotundus) blood meal shotgun-sequenced samples using a two-step metagenomics approach. In the first BLAST-alignment step, metagenomic reads were mapped to COI barcodes using BLAST. Second, mtDNA assembly of prey contigs was carried out with Novoplasty using ai) seeds from COI barcode or aii) mitochondrial DNA (mtDNA) of prey identified from the BLAST-alignment step corresponding to each sample, and b) seeds from COI barcode or mtDNA of all prey species identified from the BLAST-alignment step. Some of the elements included in the figures were obtained and modified from the Integration and Application Network, University of Maryland – Center for Environmental Science (https://ian.umces.edu/symbols/), and BioRender.com. Image of tapir from Foresman (2007). Common vampire bat image credit: Megan Griffiths. |