|
||
|
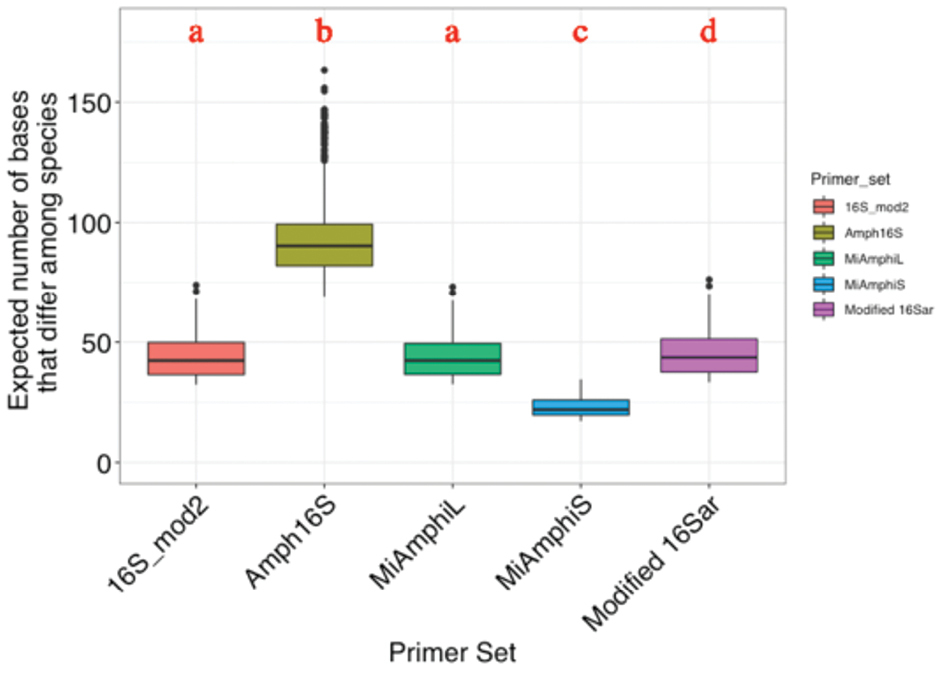
Comparison of the taxonomic resolutions amongst primer sets. The vertical axis indicates the expected number of bases that differ amongst species within the amplified region in each primer set. Each category indicates a set of primers; 16Sar_mod2: 16Sar_mod2 and Amph_16S_1070R, Amph16S: Amph_16S_1070F and Amph_16S_1340R, MiAmphiL: MiAmphiL-F and MiAmphiL-R, MiAmphiS: MiAmphiS-F and MiAmphiS-R, Modified 16Sar: Modified 16Sar and Amph_16S_1070R. The expected number of bases differed significantly amongst primer sets (ANOVA: p < 0.05). Significant differences were indicated by different letters (Tukey-Kramer test: p < 0.05). |