|
||
|
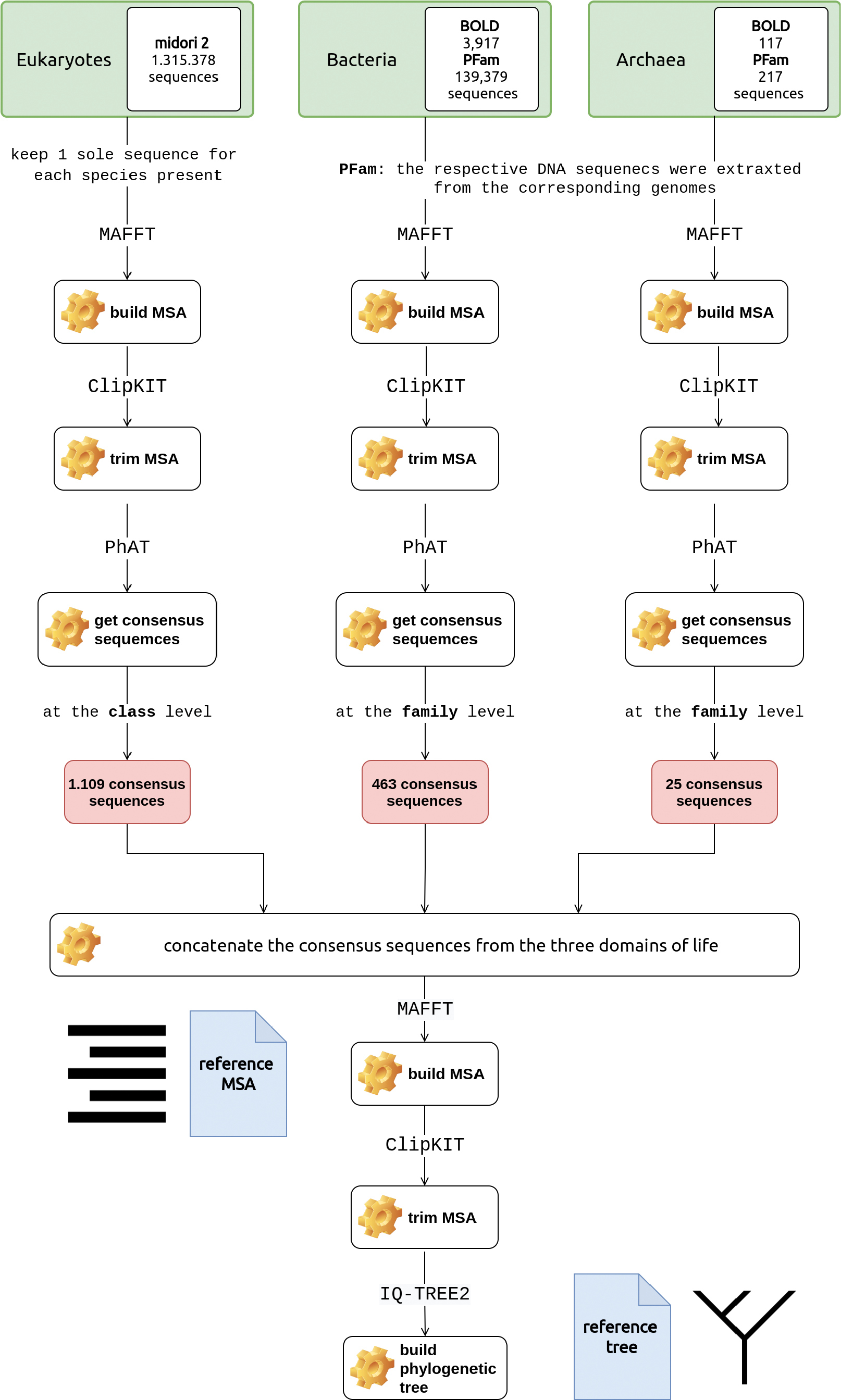
Overview of the approach followed to build the COI reference tree of life. Sequences were retrieved from Midori 2 (eukaryotes) and BOLD (bacteria and archaea) repositories. Consensus sequences at the family level were built for each domain specific dataset. MAFFT and consensus sequences at the family level were built using the PhAT algorithm. The COI reference tree was finally built using IQ-TREE2. Noun project icons by Arthur Slain and A. Beale. |