|
||
|
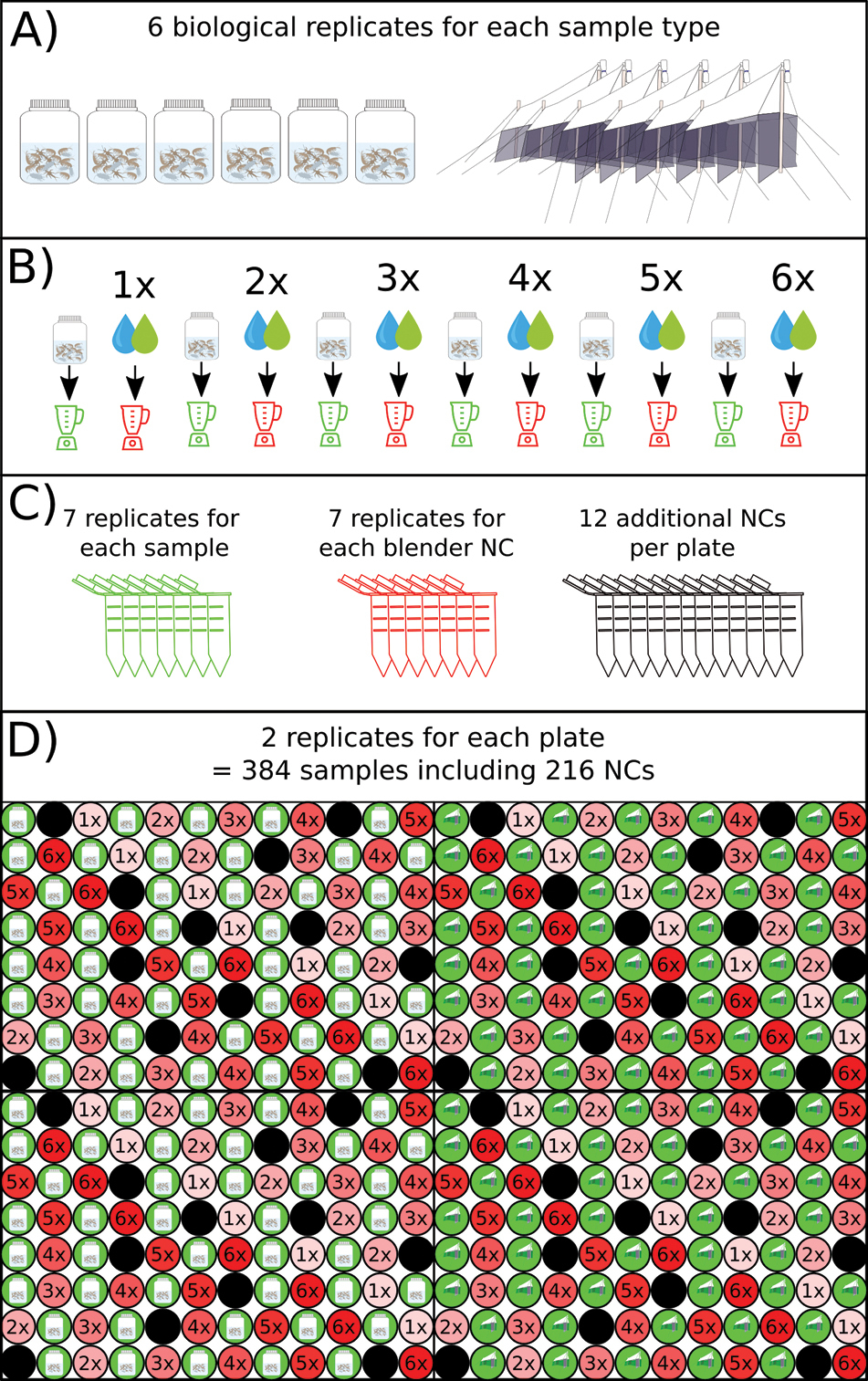
Schematic overview of the study design. A) 6 biological replicates of both sample types (stream kick-net sample, Malaise trap sample) were used in this study. B) Each of the samples were homogenized in the blender which was cleaned 1 – 6 times afterward by letting it run for 20 s with either ddH2O (blue drop, kick-net samples) or self-made decontamination solution (DIY-DS, green drop, Malaise trap samples). After cleaning, the blender was filled with EtOH to create a blender negative control. C) Each sample, as well as each blender negative control, was replicated 7 (extraction replicates) times in 2 ml tubes. At this stage, 12 additional tubes were added that never had contact with the blender to be able to distinguish possible contamination from the sample homogenisation from contamination that occurred in the downstream analysis. D) The samples were then transferred into two 96-well plates, which were replicated once more (technical replicates), to distinguish between contaminations that might have happened at stage C) from contamination that might have happened after stage D). |