|
||
|
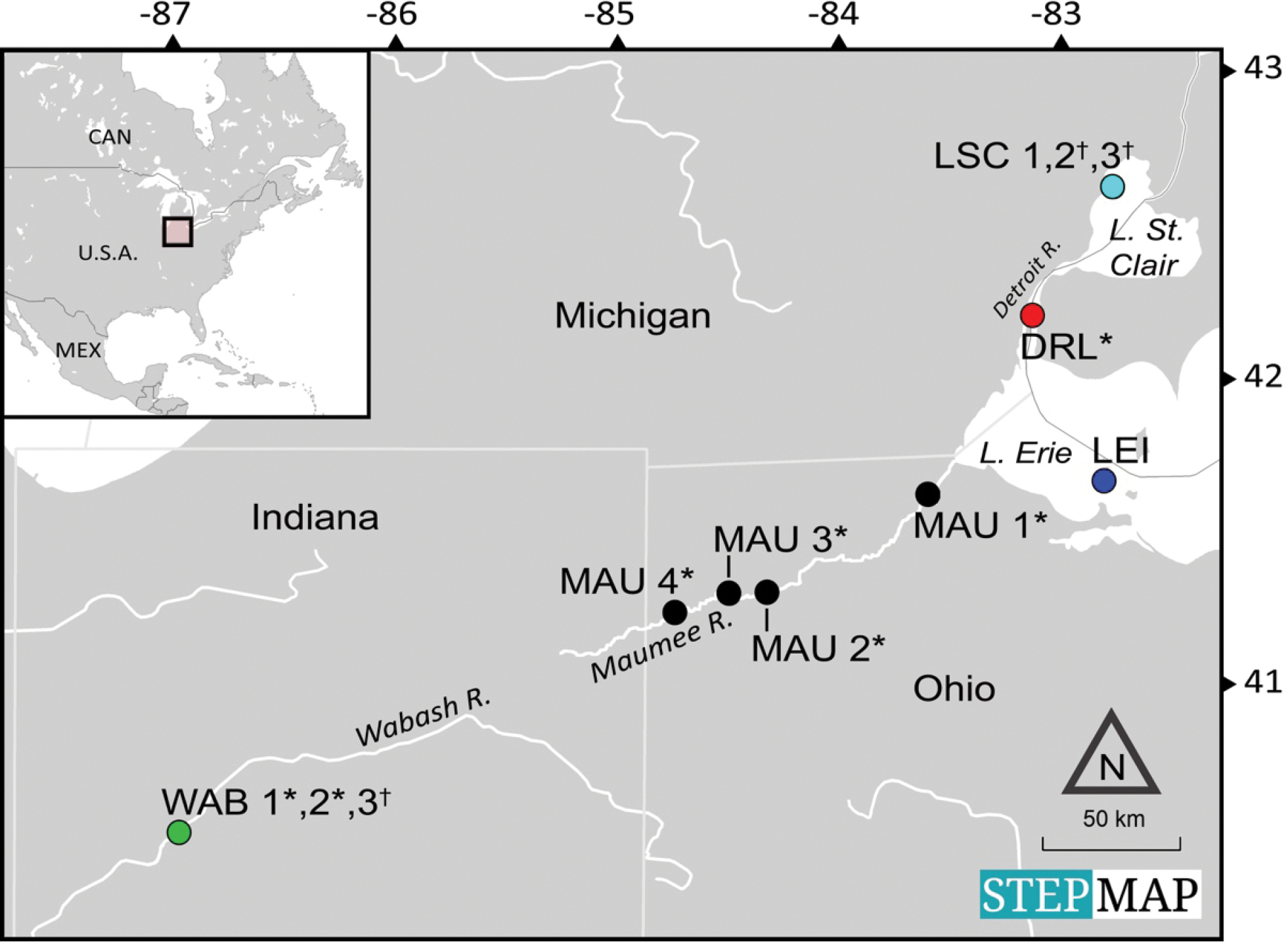
Map showing sample sites in the Wabash River (WAB), Maumee River (MAU), Detroit River (larval fish sample; DRL), Lake St. Clair (LSC), and Lake Erie Islands (LEI) (for Experiment Series A and B). At selected sites, morphological surveys (*) or traditional population genetics sampling and data collection (†) were conducted and compared to eDNA metabarcoding assay results. Wabash River (WAB) and Lake St. Clair (LSC) locations were in too close proximity to be depicted separately (geographic coordinates are in Suppl. material 1: Table S1). Field locations were mapped using STEPMAP (stepmap.com, which holds no copyright on data or layers presented). |