|
||
|
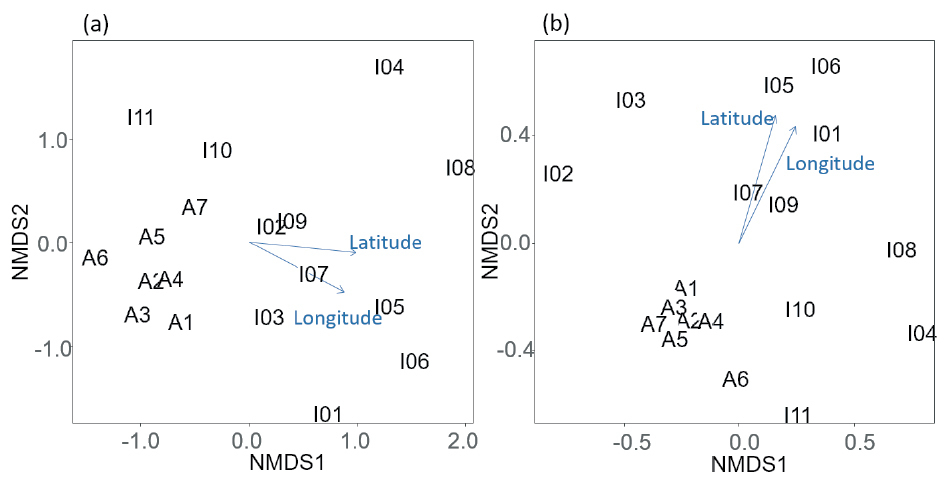
Dissimilarity of the DNA assemblages among sites as revealed by nonmetric multidimensional scaling (NMDS) ordination for (a) datasets with only fungal OTUs (stress value = 0.159) and (b) datasets using the read number (stress value = 0.153). Numbers are consistent with site numbers in Fig. 1 in the main text. The ordinations were significantly correlated with latitude and longitude for both datasets (‘envfit’ function; latitude, (a) r2 = 0.749, P = 0.0002, (b) r2 = 0.684, P = 0.0006; longitude, (a) r2 = 0.7931, P = 0.0002, (b) r2 = 0.802, P = 0.0002). |