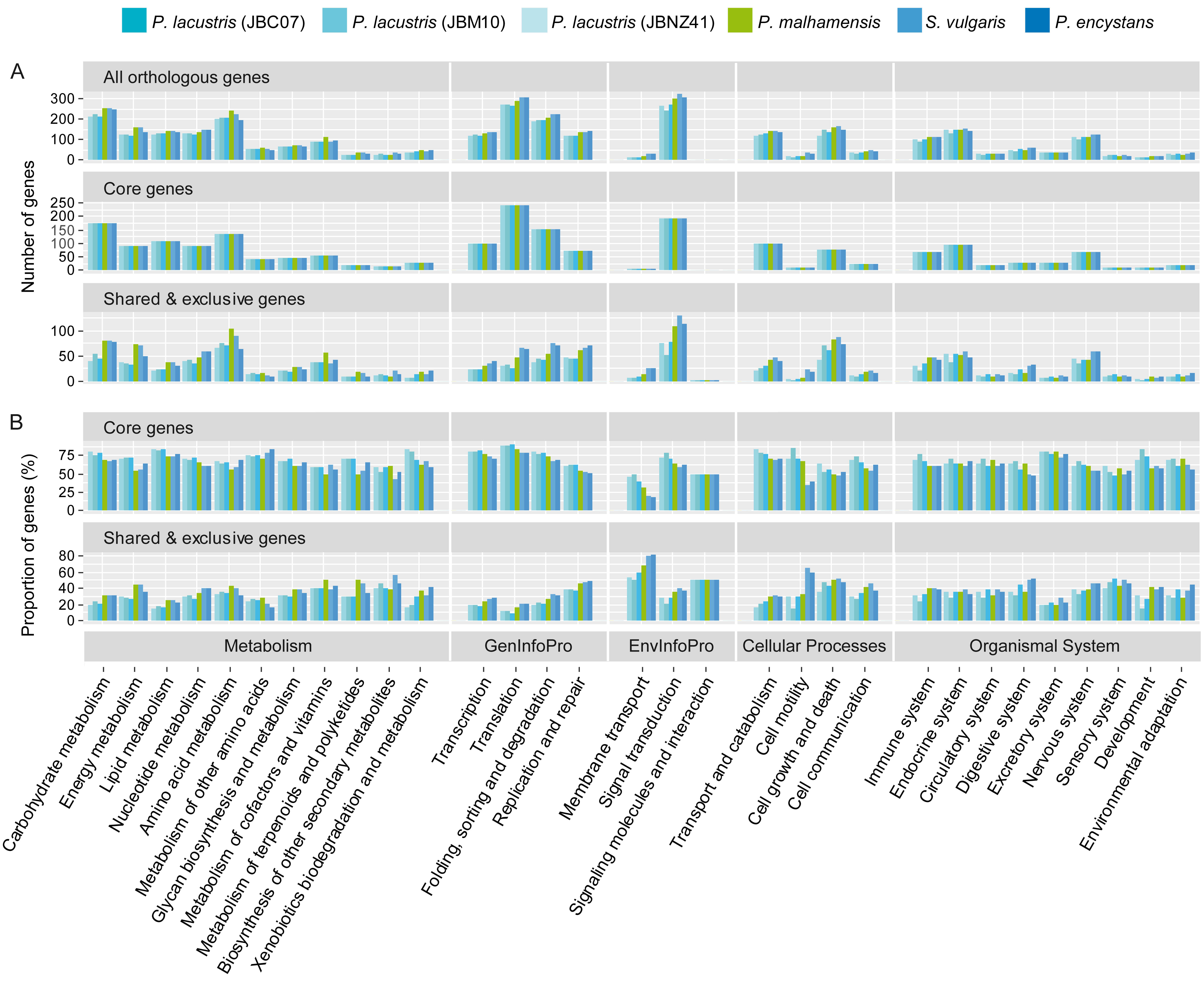
Functional assignment of non-redundant KEGG orthologous genes of Poteriospumella lacustris (JBC07, JBM10, JBNZ41), Poterioochromonas malhamensis (DS), Spumella vulgaris (199hm) and Pedospumella encystans (JBMS11) to 31 pathway groups of the KEGG categories “Metabolism”, “Genetic Information Processing” (GenInfoPro), “Environmental Information Processing” (EnvInfoPro), “Cellular Processes” and “Organismal Systems”. A All functional hits per pathway group and strain were summarized for the whole transcriptomes, for the core transcriptome of all six strains and for the shared and exclusive genes. B The percentage proportion of genes per pathway group and strain was calculated for the core genes and the shared and exclusive genes.